DSMC and PICMC documentation
2022-02-17
3.1 Parametrized postprocessing with RIG-VM
The 3D postprocessing files (*.pos) produced in simulation runs can be opened and further processed with GMSH; however, the handling of many files and performance of post processing operations such as time-averaging, cut-planes, line profiles etc. can be cumbersome this way. For that purpose, a RIG-VM module module_ppp.so1 is provided. This module offers the following:
- Importing binary post-processing (
*.pos) - files generated during DSMC / PIC-MC runs - Further arithmetic processing of imported data
- Perform time averaging
- Creation of user-specified reduced views such as cut-planes, line profiles etc.
- ASCII export of processed data
3.1.1 Prerequisites / installation
The file module_ppp.so is a module of the scripting language RIG-VM which is part of the PIC-MC software package. Within the installation directory, the module file is located in the .../lib/* directory, while the RIG-VM script interpreter is located in .../bin/*. In order to use that module from the Linux commandline, please make sure that the following environmental variables are set:
- LD_LIBRARY_PATH=$LD_LIBRARY_PATH:/path/to/picmc/lib
- PATH=$PATH:/path/to/picmc/bin
3.1.2 General usage
The postprocessing module is used within RIG-VM scripts (ASCII files, which typically have the file extension *.r). These scripts should in general be in the following format:
compiletime load "module_ppp.so"; # Import post processing module
case = BATCH.file; # Get filename of respective *.par file
PPP obj = {}; # Initiate post processing object 'obj'
# do some postprocessing stuff with obj.
# ...Assumed that the script is named extract.r, its invocation from the command line works via
rvm -b extract.r simulationcase.parIt is also possible to use the same script on multiple *.par files in batch mode:
rvm -b extract.r *.parThe following sections give a summary of all commands integrated in the postprocessing module as well as a general description of the concept. Afterwards, the functionality of the respective functions is shown in examples.
3.1.3 Summary of commands
After having initiated a post processing object obj (any other name is possible, too) as described in section (sec:ppp_rvm_generalusage?), its internal methods can be invoked as member functions.
In the following all postprocessing commands are summarized:
3.1.3.1 Functions to import data
There are three main functions for importing data:
obj.read("input.pos", "name");
Reads an input file in GMSH postprocessing format*.posand stores it under the internal namename. If multiple files are loaded under the same name, they are automatically internally averaged. This feature is mostly being used for time-averaging over multiple data snapshots taken at different times.obj.merge("input.pos", "name");
This command is used for combining multiple postprocessing files with different data coordinates into a single dataset. It is mostly being used for combining multiple files with e.g. individual particle positions taken at different timesteps into a trajectory plot.obj.append("input.pos", "name");
The append command is used to append multiple postprocessing files into a time series.
3.1.3.2 Simple cut planes
A simple functionality, which is most frequently used, is to make a 2D cutplane from a 3D dataset, which is parallel to the coordinate axes. There are the following three options for that:
obj.xy_plane(z0, delta_z, "method");
Cutplane parallel to the X-Y axes at z coordinatez0within a range for data points of \(z0\pm \delta_z\)obj.xz_plane(y0, delta_y, "method");
Cutplane parallel to the X-Z axes at y coordinatey0within a range for data points of \(y0\pm \delta_y\)obj.yz_plane(x0, delta_x, "method");
Cutplane parallel to the Y-Z axes at x coordinatex0within a range for data points of \(x0\pm \delta_x\)
The "method" parameter describes how to project data points onto the cut plane. There are three options:
"nearest_neighbour"
The nearest datapoint located in perpendicular direction within the search range is projected onto the cutplane."average"All data points located in perpendicular direction within the search range are averaged, and the average value is taken at the position of the cutplane."interpolate"The value is calculated by linear interpolation between two data points located on both sides of the cutplane within the search radius
3.1.3.3 General oblique planes and lineprofiles
Oblique cutplanes or can be created with the more general methods
obj.plane(x0, y0, z0, x1, y1, z1, x2, y2, z2, delta, N, M, "method");
obj.line( x0, y0, z0, x1, y1, z1, delta, N, "method");For a detailed description, please refer to this example.
3.1.3.4 Cylindrical cutplanes
2D cutplanes along a cylindrical surface can be obtained via
obj.cylinder(x0, y0, z0, x1, y1, z1, r_cyl, delta, nl, nphi, "method");Please refer to the example on cylindrical cutplanes for further explanation.
3.1.3.5 Output functions
The postprocessing module can create output files either in GMSH postprocessing format *.pos, Techplot format or VTK format. Depending on the type of data, the outputs can be either in scalar or vector format. For GMSH postprocessing files, there are the following options:
obj.write_pos_scalar("output.pos", "View title", expression);
obj.write_pos_vector("output.pos", "View title",
expression_x, expression_y, expression_z);
obj.write_pos_scalar_binary("output.pos", "View title", expression);
obj.write_pos_vector_binary("output.pos", "View title",
expression_x, expression_y, expression_z);
obj.write_pos_triangulate("output.pos", "View title", expression);The first two commands produce GMSH *.pos files in ASCII format, which contain either scalar or vector data. The next two commands do the same but produce files in binary *.pos format, which is somewhat compressed in comparison to the ASCII format. The last command works only after a 2D cutplane operation and produces a triangulated view of scalar data, which means that they are represented as a triangulated surface.
Each command takes the name of the outputfile and the caption of the view ("View title") as arguments, followed by one or more arithmetic expressions. The expression parameters contain an arithmetic expression, which should be evaluated at the respective datapoints of the last operation (cutplane, lineprofile etc.). They could use the variables x, y, z, which represent the actual coordinates, and in case of cylindrical cutplanes also the variable phi, which represents the azimuth angle.
Additionally, it can contain any of the variable names specified under name in the import functions as well as their maximum and minimum value via name_max and name_min. If the imported quantity is a vector dataset, the individual components are accessible via name_x, name_y, name_z, and thereof, the maxima / minima are again accessible via name_x_max, name_x_min etc.
There is an additional category of output functions, where also the coordinates of the output data can be explicitely manipulated:
obj.write_pos_scalar_parametric("output.pos", "View title",
x, y, z, expression);
obj.write_pos_scalar_binary_parametric("output.pos", "View title",
x, y, z, expression);
obj.write_pos_vector_parametric("output.pos", "View title",
x, y, z,
expression_x, expression_y, expression_z);
obj.write_pos_vector_binary_parametric("output.pos", "View title",
x, y, z,
expression_x, expression_y, expression_z);These functions work in the same way as the functions described above, but take the coordinates x, y, z as additional input. The purpose is to transform the coordinates of the output data in order to e.g. give them a coordinate offset or to project them onto a cylindrical surface.
Output functions of scalar and vector data in Techplot and VTK format work in the sam way, their syntax representations are
obj.write_tec_scalar("output.dat", "View title", expression);
obj.write_tec_vector("output.dat", "View title",
expression_x, expression_y, expression_z);
obj.write_vtk_scalar("output.vtp", "View title", expression);
obj.write_vtk_vector("output.vtp", "View title",
expression_x, expression_y, expression_z);For export of data in ASCII files (either *.txt or *.csv) the following functions are available:
obj.write_txt("output.txt",
["col1", "col2", ...], [exp1, exp2, ...]);
obj.write_csv("output.csv",
["col1", "col2", ...], [exp1, exp2, ...]);Besides the name of the output file, these functions expect a list of column names and a list of expressions for the columns (obviously, both lists have to be of the same length). The expressions in the expression list can use the same variable names as described above.
Finally, there are output functions for time series (animations). In contrast to the previous output functions, they work not (yet) with arithmetic expressions but only with variable names, as were used in the "name" argument of the append command. If needed, additional input data sets can be computed via the commands create_scalar, create_vector as described in the data manipulation section.
obj.write_pos_time_steps_scalar("output.pos",
"View title", "name");
obj.write_pos_time_steps_scalar_binary("output.pos",
"View title", "name");
obj.write_pos_time_steps_vector("output.pos",
"View title", "name");
obj.write_pos_time_steps_vector_binary("output.pos",
"View title", "name");3.1.4 Manipulation of data
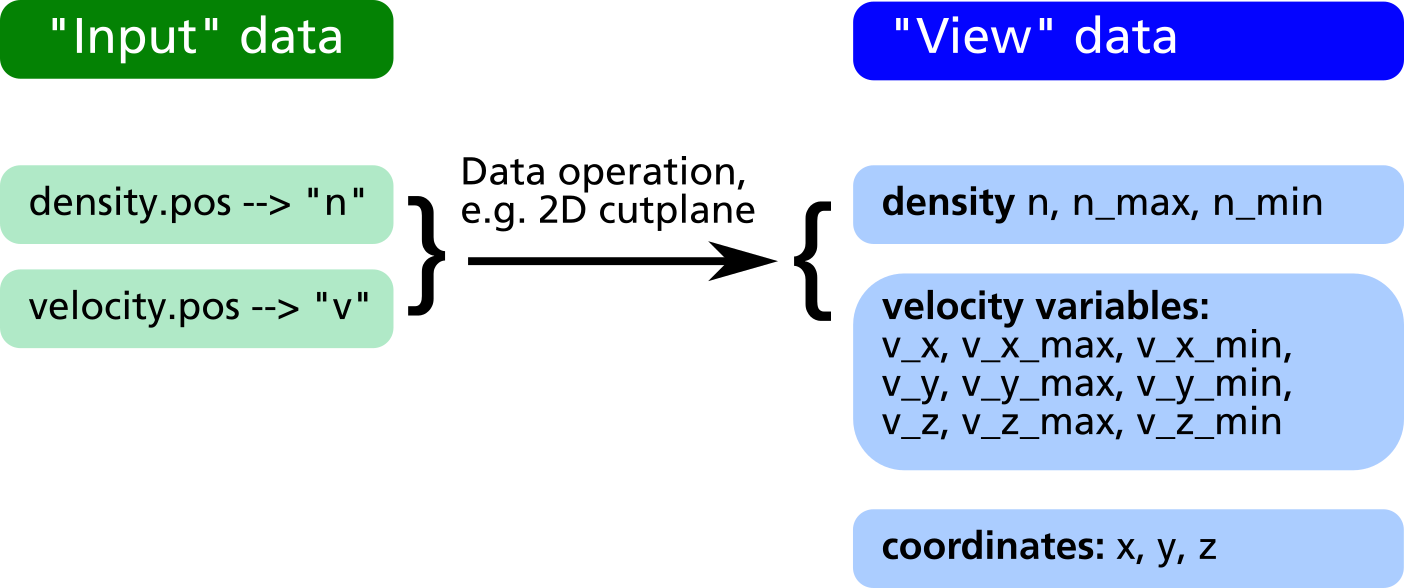
The RVM postprocessing module generally uses two different data sets, namely the "input" and the "view" data as shown in the figure below.
- The "input" data consists of the raw data obtained from the simulation output files (GMSH POS format). With every
readcommand, a new entry in the input data is created. Each input data can be assigned a short name which will be later used as variable name (e.g. "n" for density in the figure below). - The "view" data are obtained from the "input" data by some geometric transformation, e.g. a 2D cut plane or a line scan. They have the same names as the input data with some exceptions:
- If the input data are vectors (e.g. the velocity field "v" in the example below) three different "view" variables
v_x, v_y, v_zare generated in order to allow a component-wise access to the vector field. - Additional variables such as the coordinates
x, y, zas well as maximum and minimum values for each "view" variable (n_min, n_max, v_x_min, v_x_max etc.) are automatically generated and appended to the "view" dataset.
- If the input data are vectors (e.g. the velocity field "v" in the example below) three different "view" variables
- If another geometrical operation is applied, the former "view" data is being deleted and replaced by data generated from the "input" in connection with the geometrical operation.
- If no geometrical operation is applied at all, the "view" data will be a copy of all input data (in former versions, the command
obj.identity();had to be explicitely invoked for that purpose, nowadays this is automatically done internally).
After applying the geometrical operations, in the obj.write_XXX output commands, all variable names in the "view" data can be used for forming mathematical expressions defining what to extract into the output files. For the above example it is e.g. possible to extract the particle flux, i.e. the product of density and velocity, in direction of the Z coordinate as well as the relative density distribution in %:
compiletime load "module_ppp.so";
PPP obj = {};
obj.read("density.pos", "n");
obj.read("velocity.pos", "v");
obj.xy_plane(0, 10, "nearest_neighbour");
obj.write_pos_scalar_binary("flux-Z-2D.pos", "Vertical flux [1/m^2 s]", n*v_z);
obj.write_pos_scalar_binary("density-relative.pos", "Relative density [1/m^2 s]",
100*n/n_max);3.1.4.1 Creation of new input data
Usually, the input data is obtained directly via the command obj.read("filename", "label");. In some cases it may be useful to create additional input data via an algebraic formula from the existing data. For this purpose, the following commands can be used:
obj.create_scalar("new_label", expression);
Create new scalar input data entryobj.create_vector("new_label", exp_x, exp_y, exp_z);
Create new vector input data entry
In the example above, it is possible to plot the relative density distribution in % because of the existence of the view variable n_max. However it is not clear how to plot e.g. the relative flux distribution, since there is no obvious way to obtain the maximum value of n*v_z. By creation of a "flux" variable as new input entry, this problem can be solved (see example listing and data structure diagram below).
compiletime load "module_ppp.so";
PPP obj = {};
obj.read("density.pos", "n");
obj.read("velocity.pos", "v");
obj.create_vector("flux", n*v_x, n*v_y, n*v_z);
obj.xy_plane(0, 10, "nearest_neighbour");
obj.write_pos_scalar_binary("flux-Z.pos",
"Vertical flux [1/m^2 s]", flux_z);
obj.write_pos_scalar_binary("flux-Z-relative.pos",
"Relative Vertical flux [%]",
100*flux_z/flux_z_max);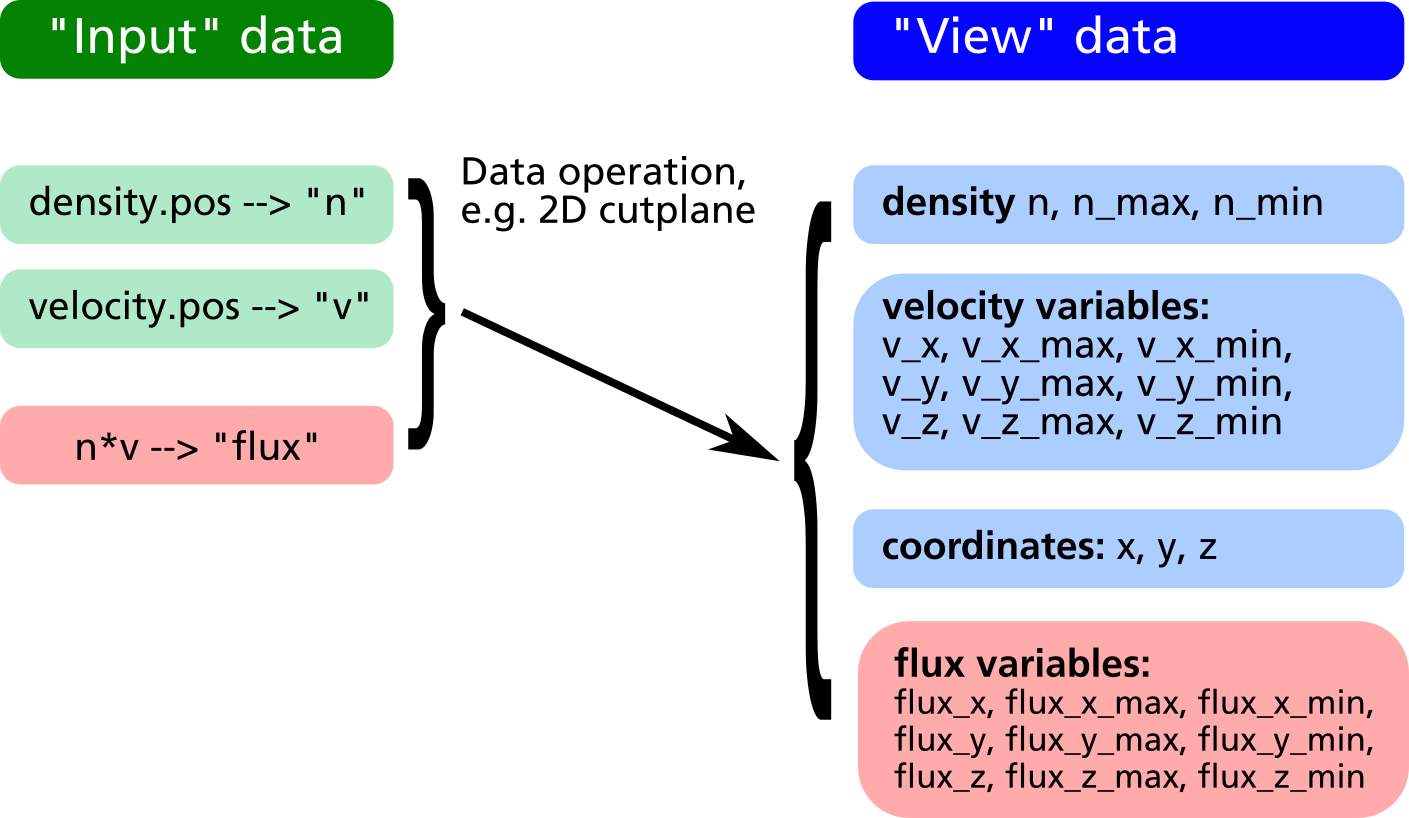
3.1.4.2 Reduction of input data
Let us assume we have 3D data of the density and velocity of evaporated atoms. In the XY-plane at Z=100 there is a substrate where the evaporated atoms can condensate.
First, in order to get the thickness profile on the substrate, the flux has to be extracted by multiplying density and z component of velocity in a cut-plane just below the substrate surface.
Second, let us assume the substrate is moving into Y direction, i.e. we have a dynamic deposition. In such cases, only the linescan of the profile in X direction averaged over Y is of relevance.
In order to do this, in prior versions of the post processing module it was required to make the 2D cut plane first, save it into a temporary POS file, load this POS file into a second post processing object and finally extract the line profile from there.
From version 2.4.3 on, there is the possibility of reducing the input data according to the last executed geometric operation, the command syntax is:
obj.reduce();
Reduction of input data space according to current arrangement of view data.
This allows for obtaining a line profile from 3D data in a much easier way:
compiletime load "module_ppp.so";
PPP obj = {};
aperture_width = 100; # Aperture width on substrate in transport direction
obj.read("density.pos", "n");
obj.read("velocity.pos", "v");
# Define vertical flux component
obj.create_scalar("Fz", n*v_z);
# Make cut plane of data just below the substrate surface
obj.xy_plane(99.5, 10, "nearest_neighbour");
# Reduce input data to cut-plane (this is the important command)
obj.reduce();
# Make another cut plane averaging over the aperture_width
# in order to get the line profile
obj.xz_plane(0, aperture_width/2, "average");
# Write line profile into TXT file
obj.write_txt("absorption_line.txt",
["x", "avg_absorption", "rel_absorption"],
[x, Fz, 100*Fz/Fz_max]);3.1.4.3 Control of written data
Regarding the format of written data, there are the following possibilities:
- The coordinates, where the written data get plotted in POS files, can be explicitely specified
- It is possible to set a flag deciding whether to write data at a given coordinate point or not
The first option involves the following output methods:
obj.write_pos_scalar_parametric("filename.pos", "Legend", x, y, z, expression);obj.write_pos_scalar_binary_parametric("filename.pos", "Legend", x, y, z, expression);obj.write_pos_vector_parametric("filename.pos", "Legend", x, y, z, exp_x, exp_y, exp_z);obj.write_pos_vector_binary_parametric("filename.pos", "Legend", x, y, z, exp_x, exp_y, exp_z);
The first two commands are for parametrized scalar plots (in ASCII and binary format, respectively), the second two commands are for vector plots. An example is given in the following listing.
compiletime load "module_ppp.so";
PPP obj = {};
obj.read("density.pos", "n");
obj.read("velocity.pos", "v");
obj.xy_plane(100, 10, "nearest_neighbour");
# Plot density and flux side by side. A coordinate offset allows for
# loading both pos files into GMSH and have the cut planes non-overlapping
obj.write_pos_scalar_binary_parametric("density-2D.pos", "Density [1/m^3]",
x, y, z, n);
obj.write_pos_scalar_binary_parametric("flux-2D.pos", "Flux [1/m^3]",
x+200, y, z, n*v_z);In order to control explicitely, which data point should be written and which should be omitted, there is the possibility to define a write_flag variable within the postprocessing object. This is demonstrated in the following:
compiletime load "module_ppp.so";
PPP obj = {
write_flag: A>0;
};
obj.read("absorption.pos", "A");
obj.write_pos_scalar_binary("absorption-noZeros.pos", "Absorption [1/m^2 s]", A);In a large 3D absorption file, non-zero values only occur at the intersection between volume grid and mesh elements of walls. Thus a large fraction of the POS file consists of zeros, which is a waste in memory consumption. With the above script, the write_flag variable forces the write routine to only output those values with non-zero absorption, i.e. A>0.
It is important to define write_flag with a colon and not to write write_flag = (A>0);. In the rvm scripting language, the colon means that the value of write_flag will be only computed on request according to the currently defined parameters on right side. With the regular "=" assignment operator, the value of the variable A will be required at the moment of the definition of write_flag, which is not the case in the above script.
It is also possible to combine multiple conditions e.g. in order to specify a certain coordinate range for output:
compiletime load "module_ppp.so";
PPP obj = {
write_flag: (abs(x)<100) and (y>-50) and (y<150);
};
# ...3.1.4.4 Convolution function
A convolution function has been implemented to operate on target absorptions plots in an attempt to model erosion profiles in MoveMag aplications. The primary approach here is to assume plasma and ion bombardment, respectively do not change with magnetron position which should be valid for small displacements vectors. Thus, we simply integrate over time the convolution of a time deviant, i.e. moving absorption plot with a stationary step function.
| Convolution and integration over time of moving source data with stationaly step function | Absorption plot (top) and convolution result (bottom) for a periodic oscillation in y |
|---|---|
 |
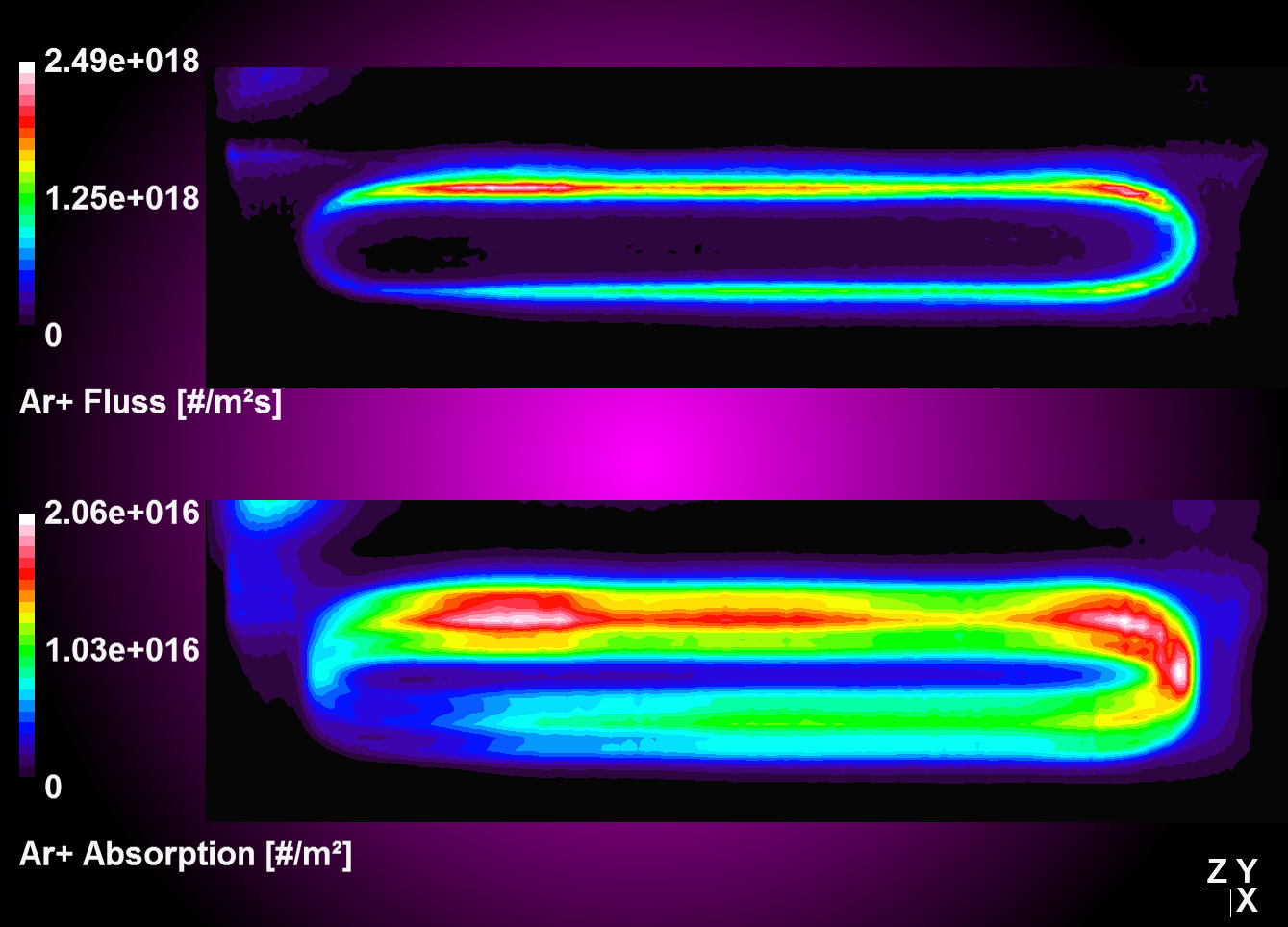 |
compiletime load "module_ppp.so";
PPP obj1 = {};
#Define velocity vector and integration time
float vx[4], vy[4], vz[4]; #[m/s]
float T = 4; #[s]
#Oscillation starting from center position
#Move 1s to left with 1 m/s, then 2 times to right accordantly, then left again.
vx[0] = -1;
vx[1] = +1;
vx[2] = +1;
vx[3] = -1;
obj1.read("absorption_Arplus_100.00us.pos", "val");
obj1.xz_plane(0.0088, 1, "nearest_neighbour");
obj1.reduce; #Operate on cut plane (2D) data set for better performance!
obj1.convolution(vx, vy, vz, T, 0);
obj1.write_pos_scalar_binary("absorption_Arplus_100.00us_convolution.pos", val);3.1.4.5 Convolution with linear weighting
Entering '1' for the weighting flag enables a linear weighting scheme during convolution operation from 1 (no displacement) to 0 (maximum displacment). With this you can realize an interpolation between two or more states, i.e. magnetron positions, by merging the results from corresponding convolution operations together.
| Convolution with constant weighting | Convolution with linear weighting |
|---|---|
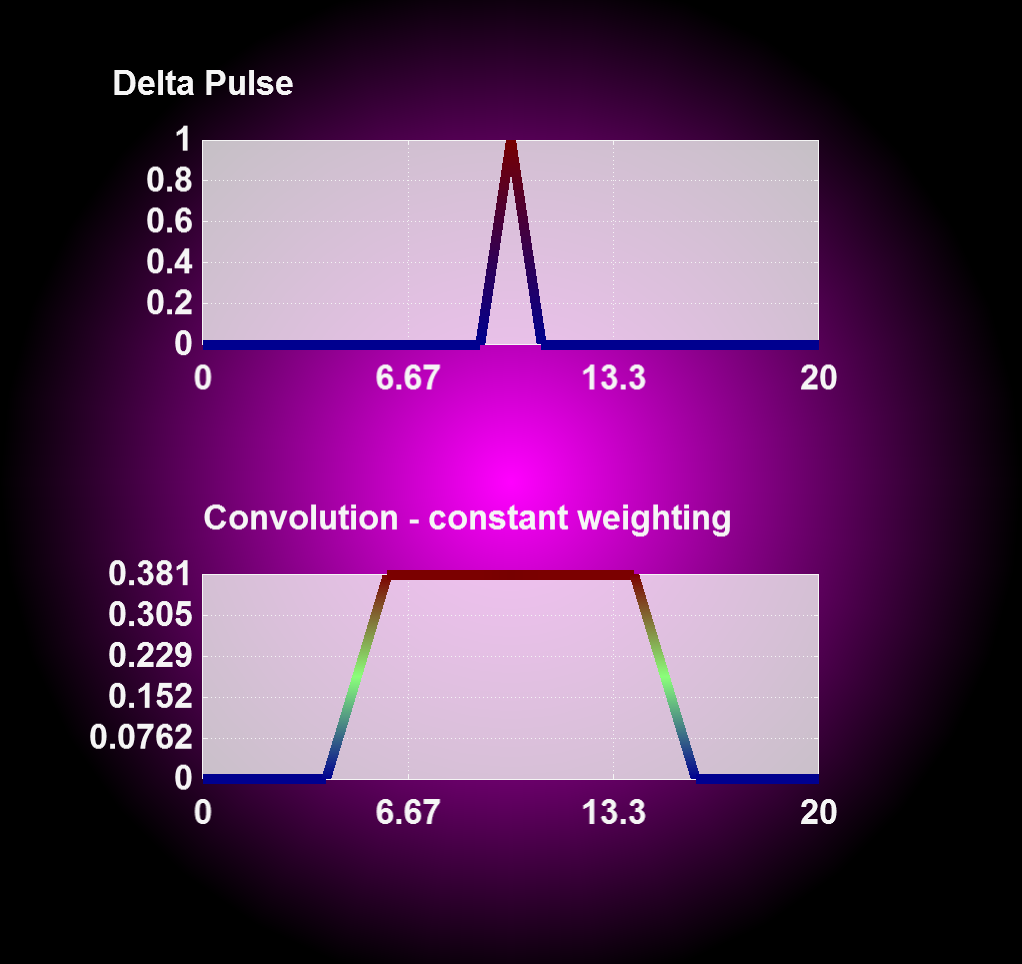 |
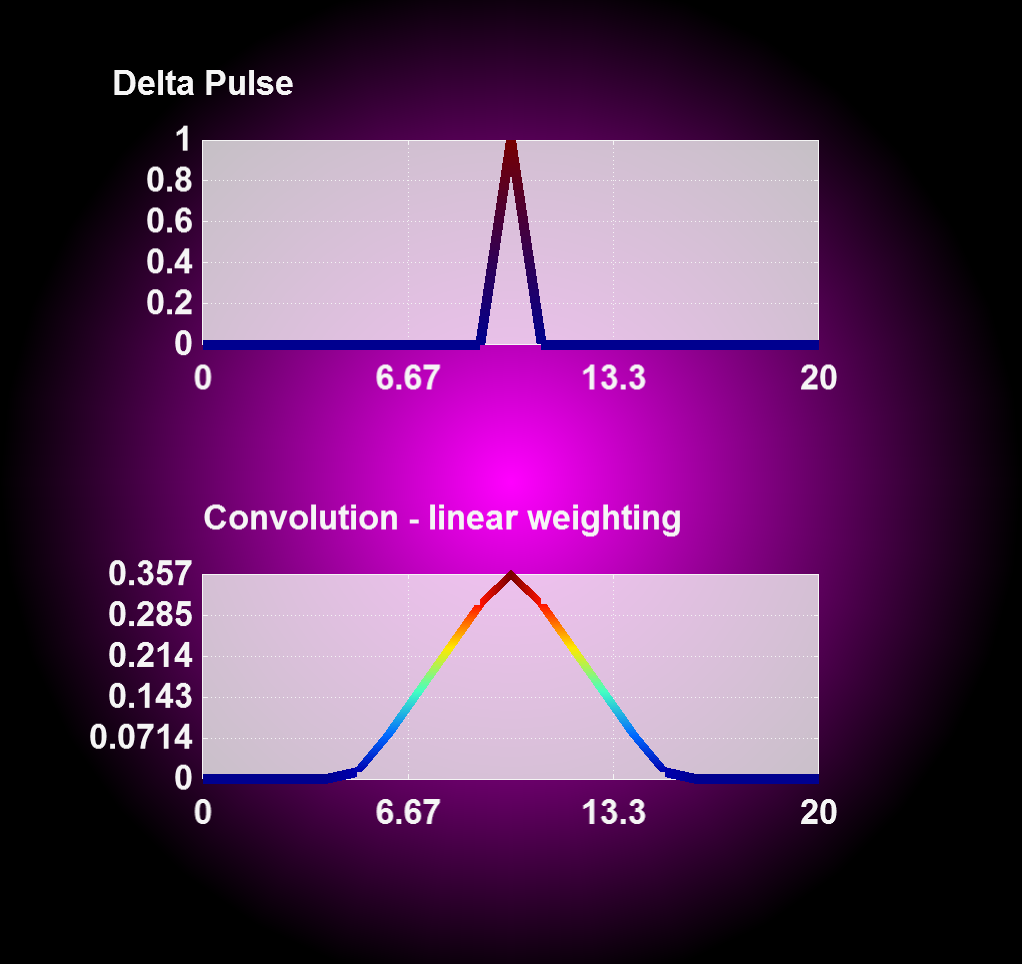 |
3.1.4.6 Circular Convolution
For a 360° convolution over the plot center use the function 'convolution_circular()' as seen below. This function has been implemented to adress modeling of absorption profiles on rotating substrates.
compiletime load "module_ppp.so";
PPP obj1 = {};
obj1.read("absorption_Arplus_100.00us.pos", "val");
obj1.xz_plane(0.0088, 1, "nearest_neighbour");
obj1.reduce; #Operate on cut plane (2D) data set for better performance!
obj1.convolution_circular();
obj1.write_pos_scalar_binary("absorption_Arplus_100.00us_convolution.pos", val);| Source data | After circular convolution |
|---|---|
 |
 |
obj.create_scalar("variable_name", expression);
obj.create_vector("variable_name", expression_x, expression_y, expression_z);
result = obj.integrate(expression);
obj.identity();
obj.reduce();
obj.convolution(vx, vy, vz, T, interpolation);
obj.convolution_circular();
The acronym PPP stands for PIC-MC Postprocessing.↩︎